How to Determine Which Enzymes to Use to Cut Plasmid
Which of the following enzymes could you use to cut the DNA SO that it has compatible sticky ends with the plasmid cut with Bglil. The basic steps are.
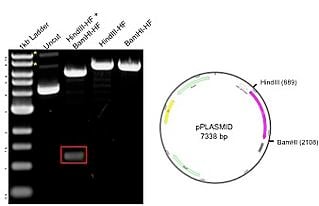
Plasmids 101 How To Verify Your Plasmid Using A Restriction Digest Analysis
However some produce blunt ends.

. Lane C Od Lane D Oe. Thaw your plasmid DNA from the minipreps performed previously. For some reason I thought that the formula needed to figure this out is.
Lane B O c. In reality this is not so problematic nowadays as most modern vectors include an artificial stretch of DNA with many different restriction endonuclease cutting sites. On the default Graphical View page you can select 1 cutters 2 cutters 3 cutters or List 0 cutters.
Grow up lots of plasmid-carrying bacteria and use them as factories to make the protein. Many restriction enzymes make staggered cuts producing ends with single-stranded DNA overhangs. 144 x 7300.
Type III cut DNA near recognition sequences. A 1agarose gel was used to run the digestions and after staining it with ethidium bromide solution the cleaved DNA fragments were visible under a trans. But it gives me such a low number that I dont think its right so Im just a little confused.
This is greatly facilitated by the use of specific restriction endonucleases. This technique is used to copy DNA that is present in small amounts such as a droplet of blood at a crime scene. I have a plasmid that is 73Kb and am told that it is cut with an enzyme of 4bp.
Insert the plasmid into bacteria. None of the above Cut with Spel Select one. PBR322 is a small cloning vector with several unique restriction sites.
This means its important to check whether plasmid and selected restriction enzymes are compatible. DNA genomic is used for PCR amplicon. The pieces backbone Csy4 LgBiTSmBiT were assembled into a fully functional plasmid with traditional subcloning techniques using unique enzyme restriction sites.
In last weeks lab you analyzed the DNA sequence of the plasmid and the plasmid with the gene of interest in order to determine how they would be cut by particular restriction enzymes. Lane B OcLane C Od Lane D O e Lane E Of. It forms the backbone of many larger more sophisticated plasmids in use today.
The plasmid is reproduced in bacteria making more copies. After completing this experiment students understood the inverse relationship between changes in FRET and changes in distance and understood how changes. None of these Cut with Acll Select one.
Otherwise the electrophoretic gels could not give any meaningful sequence. Each enzyme recognizes one or a few target sequences and cuts DNA at or near those sequences. Type II cut DNA within or close to the recognition sequence.
Buffers used in double digestions must be compatible ie both restriction enzymes should cut when combined with a particular buffer. Open the Digests button navigate to New Digest and specify NEB and Double Cutters in the settings. For a full list of REs with recognition sites within the DNA molecule select Custom Digest.
Answer 1 of 3. Restriction enzymes are DNA-cutting enzymes. You need a software to know where your enzymes of interest will cut plasmid pGFPuv.
Select enzymes of interest and then click Digest to visualize where the enzymes cut on the DNA molecule. The aim of this experiment is to determine whether the unknown plasmid DNA is cut into different sized fragments by using a variety of restriction enzymes- namely EcoR1 Pst1 and BamH1. Spel Od Bell Oe.
Consider buffer and temperature compatibility when digesting with more than one enzyme. Try choosing unique enzymes. Generally Type I enzymes cut DNA at locations distant to the recognition sequence.
DNA genomic is digested by restriction enzyme then an oligo-targeter is added for detection. O a Acil O b. DNA ligase is a DNA-joining enzyme.
Restriction analysis of Plasmid DNA In this exercise you will digest the plasmid pBR322 with 5 different restriction enzymes and resolve the fragments by agarose gel electrophoresis. DNA Restriction from D. Lane A O b.
Cut open the plasmid and paste in the gene. Watch out for methylation issues. Select restriction enzymes to digest your plasmid.
A technique which produces many copies of a target DNA sequence. This process relies on restriction enzymes which cut DNA and DNA ligase which joins DNA. Search by the number of cuts 1 find and left-click on AseI to select this enzyme and hit Run Digest.
In the following weeks we will determine the sizes of the produced DNA fragments and will use this. Like those suggested by Prof Yoram Gerchman and for the pipoetting follow manufacturers prescriptions Cite. And Type IV cleave methylated DNA.
Enzymes that only cut once allow you to more easily and accurately visualize the full size of your construct. Consult the Promega enzyme buffer chart on the last two pages of this lab to determine the type of buffer to use in the double digestions. In todays lab you will use restriction enzymes to cut the plasmid DNA you are going to purify.
This is important because it ensures that the fragments end in a particular base AGC or T. Lane A O b. To determine which restriction enzymes will cut your DNA sequence and where they will cut use a sequence analysis program such as Addgenes Sequence Analyzer.
Open and switch the view to Linear Map. Determine an appropriate reaction buffer by reading the instructions for your enzyme. Researchers take a part of DNA such as a gene and insert it into a circular DNA molecule called a plasmid.
Consult the manufacturers manual for the optimal working conditions for each enzyme.
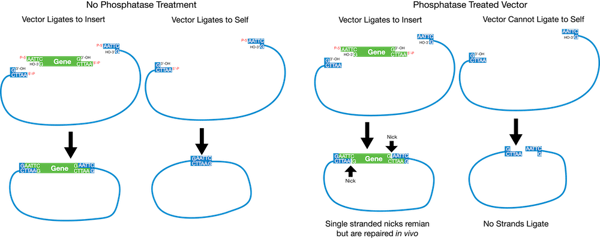
Plasmids 101 Restriction Cloning
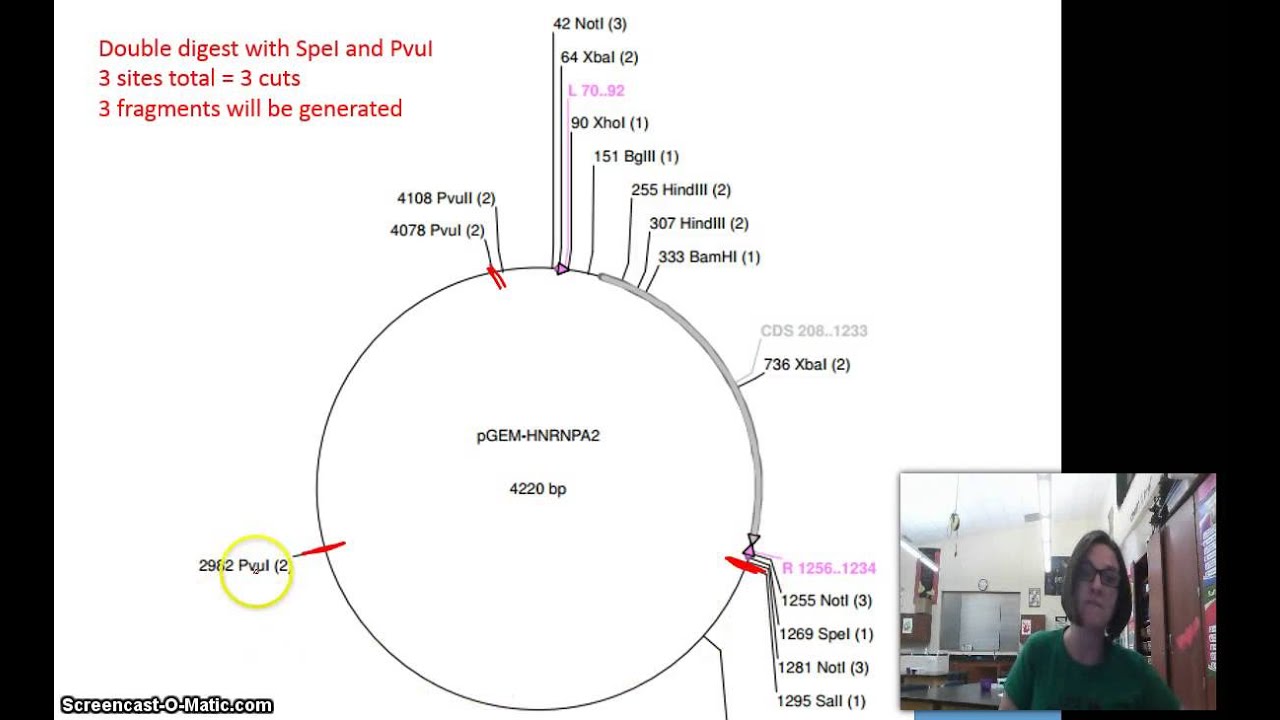
How To Read A Vector Map For A Restriction Digest Youtube

Ap Biology Restriction Enzyme Digests On Circular Plasmids Youtube
No comments for "How to Determine Which Enzymes to Use to Cut Plasmid"
Post a Comment